K-Means in Python
I’ve written a general K-Means implementation in Python. It can be found here:
https://github.com/mattnedrich/algorithms/blob/master/python/clustering/kmeans/kmeans.py
Everything is contained in one file, kmeans.py The usage model is the following
import kmeans # observations # - list of items to be clustered # numMeans # - number of clusters desired # featureVectorFunc # - function that extracts a feature vector (in the form of a Tuple) from an observation # maxIterations # - optional param, can be used to set a max number of iterations [clusters, error, numIter] = kmeans.cluster(observations, numMeans, featureVectorFunc, maxIterations)
My goal was to make something simple and accessible that doesn’t require additional libraries (e.g., numpy is not required). The user must do one thing - provide a featureVectorFunc that takes in an observation from the input, and returns a feature vector in the form of a Tuple. This function would look something like this:
def someFeatureVectorFunc(obs): featureVectorAsTuple = extractFeatureVector(obs) return featureVectorAsTuple
The output clusters is a list of cluster objects. Each cluster has a field called observations. This is subset of the original input observations (stored in a list).
Example Results
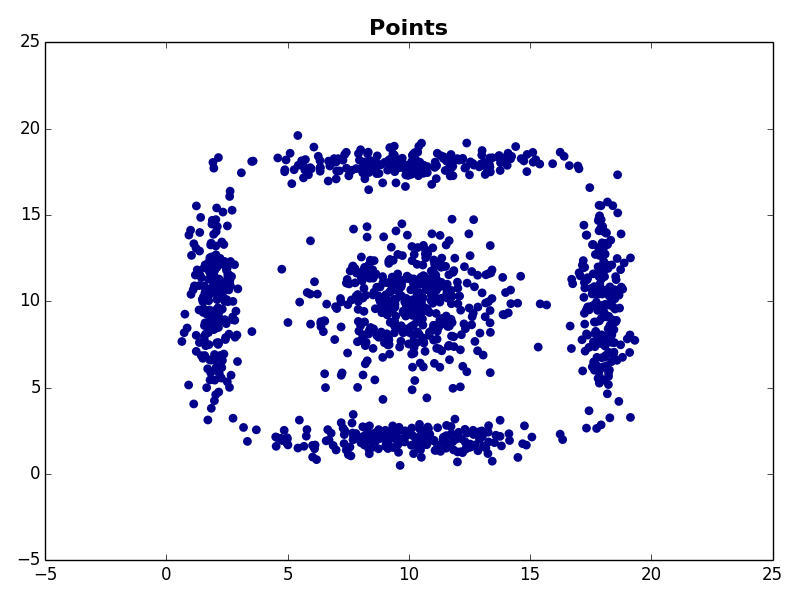
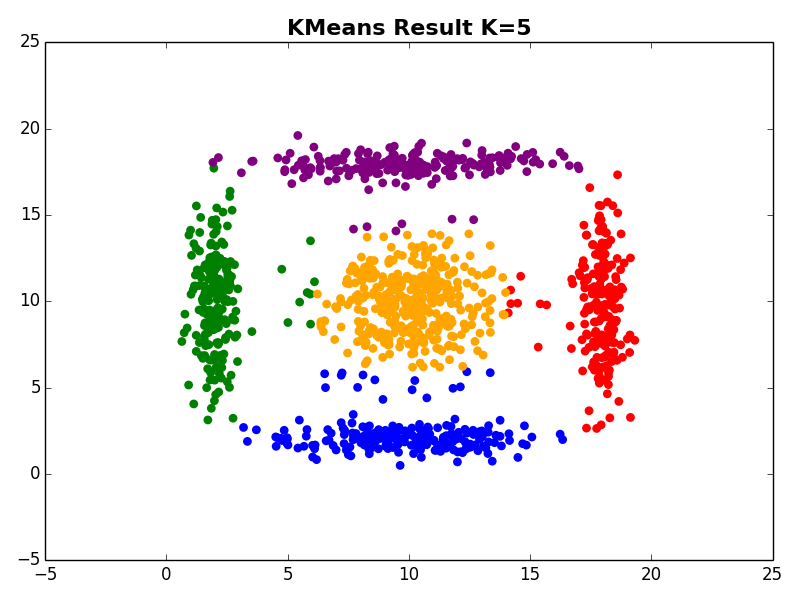
I ran my implementation on the three synthetic datasets below. For each dataset, I used K=5. I also ran K-Means 10 times and selected the result with the lowest error.
Dataset 1
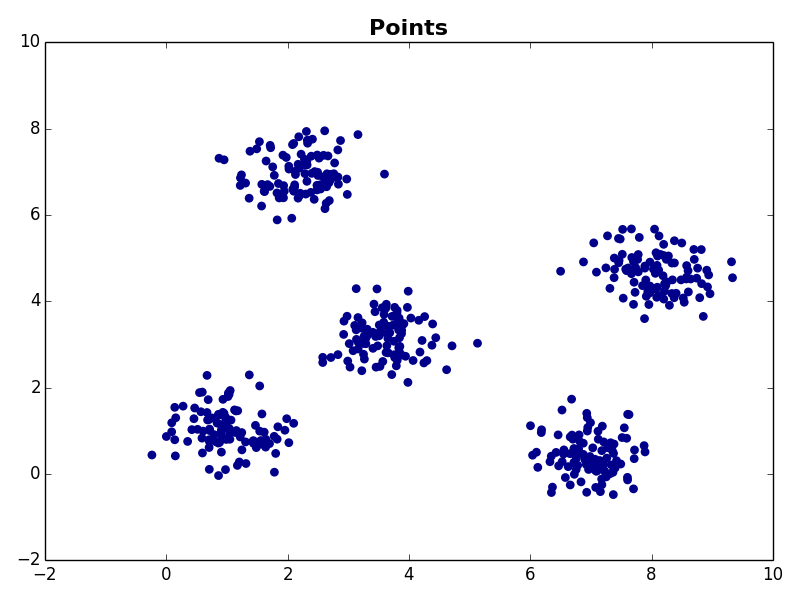
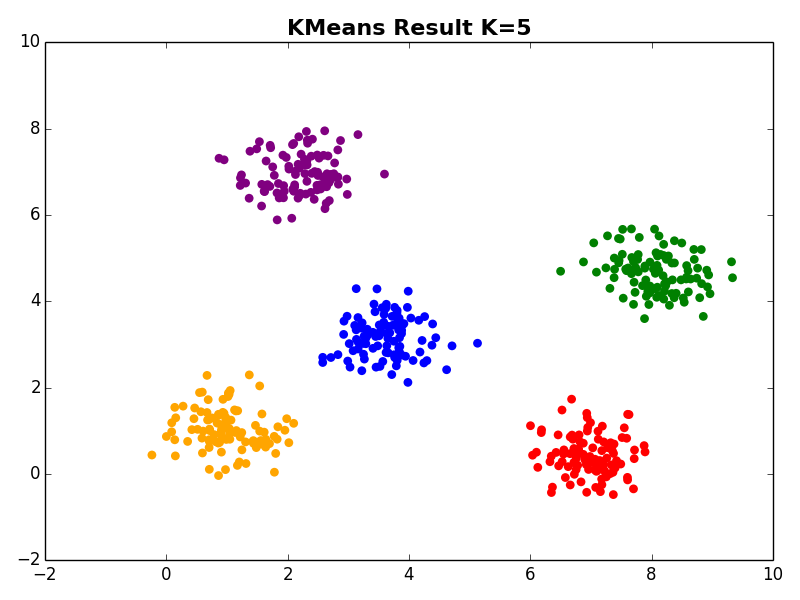
Dataset 2
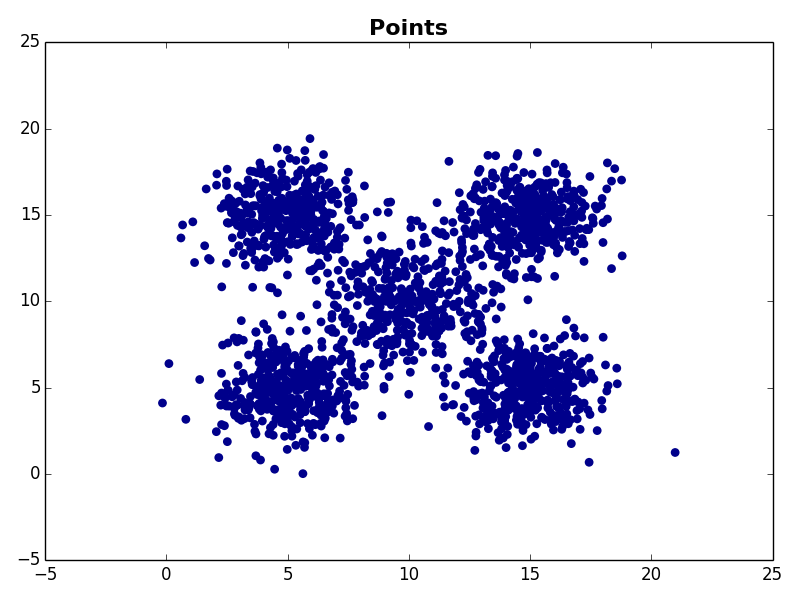
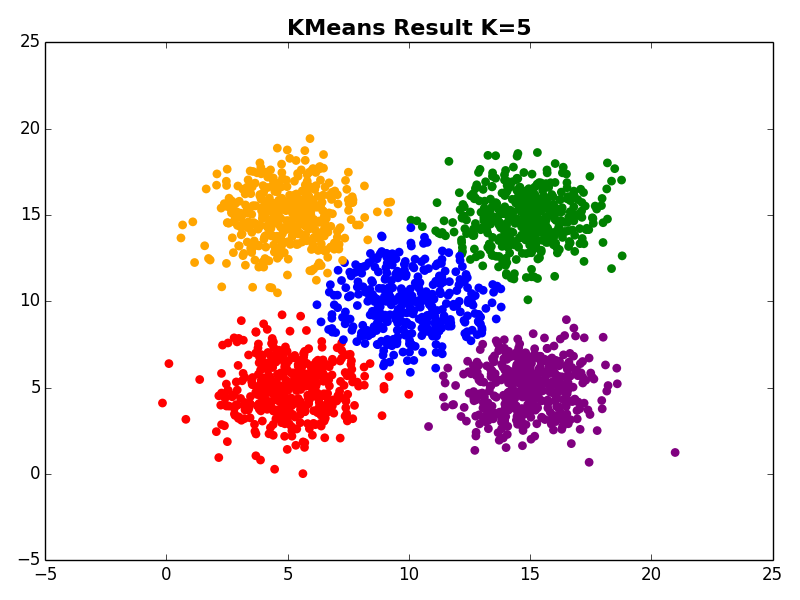
Dataset 3